Our
Research
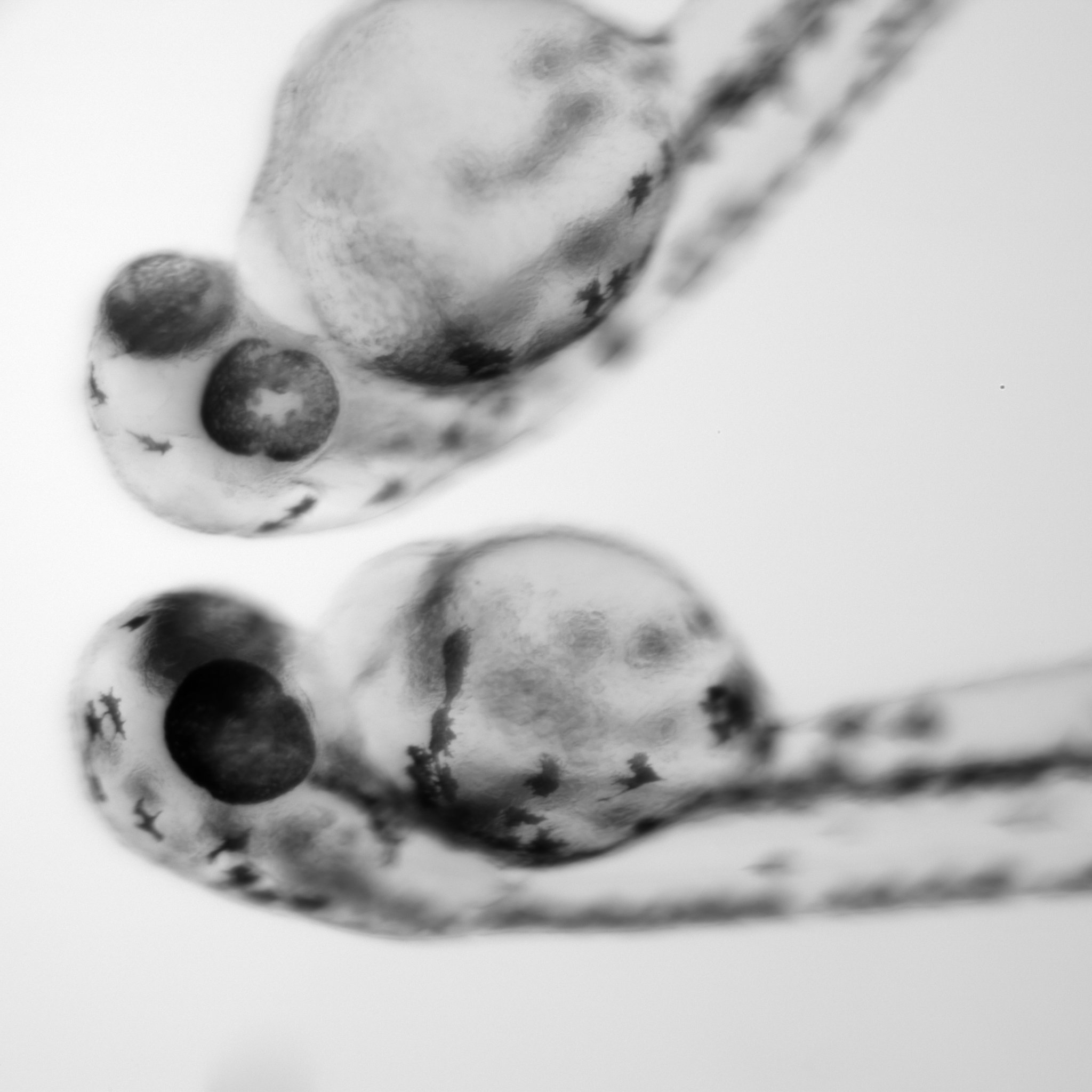
We investigate the regulation of neural cell fate decisions using the zebrafish brain as a model.
Our goal is to identify how different classes of neurons are generated from various progenitor pools.
We wish to apply the principles that we discern during development to investigate regeneration and neurodevelopmental disorders.
-
We identify how many molecularly distinct cell types exist in the developing brain and identify their timeline of appearance.
This is important to generate a “ground truth” atlas that represents diversity during normal development, and also to identify rare cell types and progenitor groups that have not been well studied.
-
We interrogate transcriptional programs that accompany and regulate neurogenesis from early to late stages of development. We use single-cell transcriptomics to obtain gene expression profiles of neural progenitors as they transition through various cell states of specification towards their terminal identities.
These studies are important to identify candidate regulators for mutant analysis.
-
Similar to analyzing transcriptional changes, we also determine changes in DNA accessibility around transcribed regions during neurogenesis from early to late stages of development. This enables us to identify regulatory elements (e.g. enhancers, binding motifs) that are important for promoting or inhibiting neuronal specification.
These studies are important to identify the hierarchy of genetic changes that mediate cell specification, and identify how this architecture may be disrupted in neurodevelopmental diseases.
-
We are studying how signaling inputs received by cells during development regulate cell fate decisions. Specifically, we are interested in identifying whether progenitor cells within a niche receive different signals and give rise to distinct types of neurons.
Signaling pathways mediate critical developmental processes including patterning and cell fate. Our goal is to derive signaling atlases that complement our transcription and chromatin maps. These studies are important to understand where and when molecular architectures of cells are disrupted in neurodevelopmental disease.
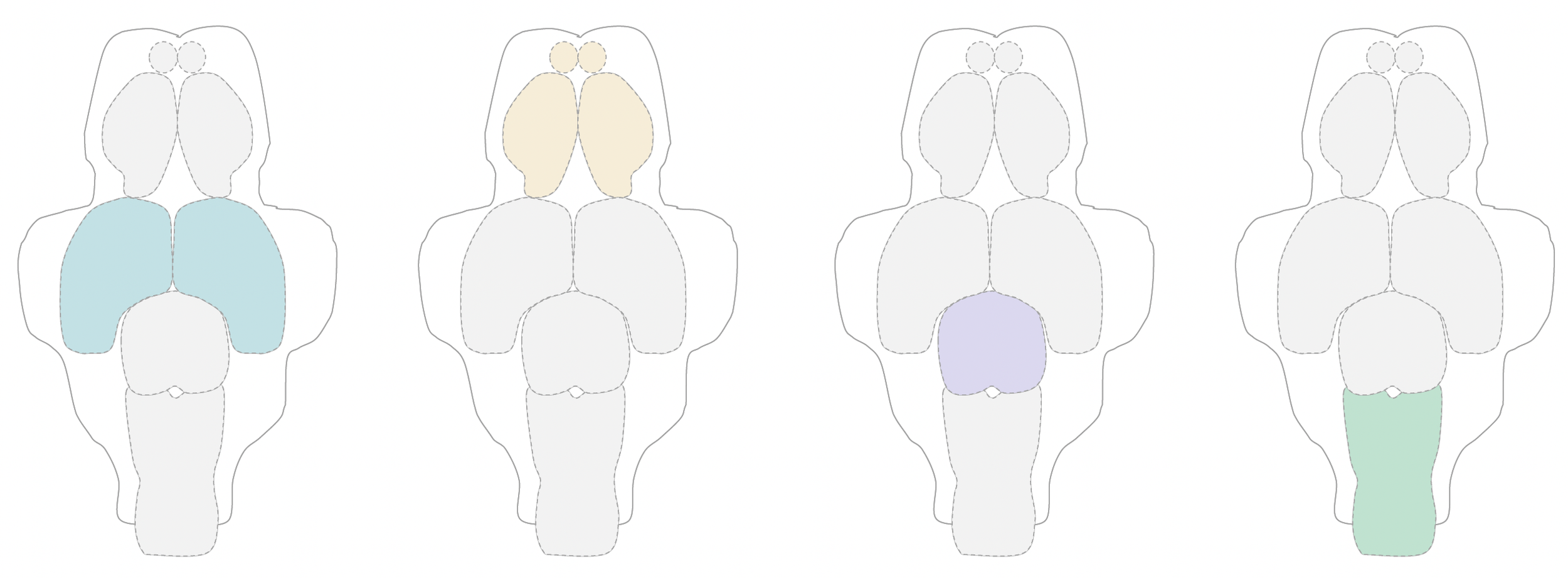
PROGENITOR HETEROGENEITY
As the brain develops, neural progenitor pools transition through many cellular states of commitment, diversification, and terminal differentiation.
Our developmental profiling data indicate that embryonic neural progenitors in the zebrafish brain are transcriptionally distinct from larval neural progenitors. These cell state changes might reflect shifts in the animal’s developmental programs that are critical for massive expansion of neuron subtypes at later stages. We are exploring these changes at the transcript and chromatin levels to address how they impact brain organogenesis.
NEURONAL DIVERSIFICATION
We have generated multiple single-cell transcriptomics datasets of the developing zebrafish brain encompassing several stages.
These data enabled comparison of cell types as the brain diversifies, and dissection of molecular trajectories that underlie cell specification and differentiation. We are using this resource to infer biological pathways towards generation of additional cell types, and testing their importance using knockout strategies. Additionally, we are overlaying multiple ‘omics datasets to generate a comprehensive map of how gene regulatory layers are coordinated during brain development, and how they become disrupted in mutants with neurodevelopmental defects.
CRISPR-Cas developmental barcoding
LINEAGE TREES
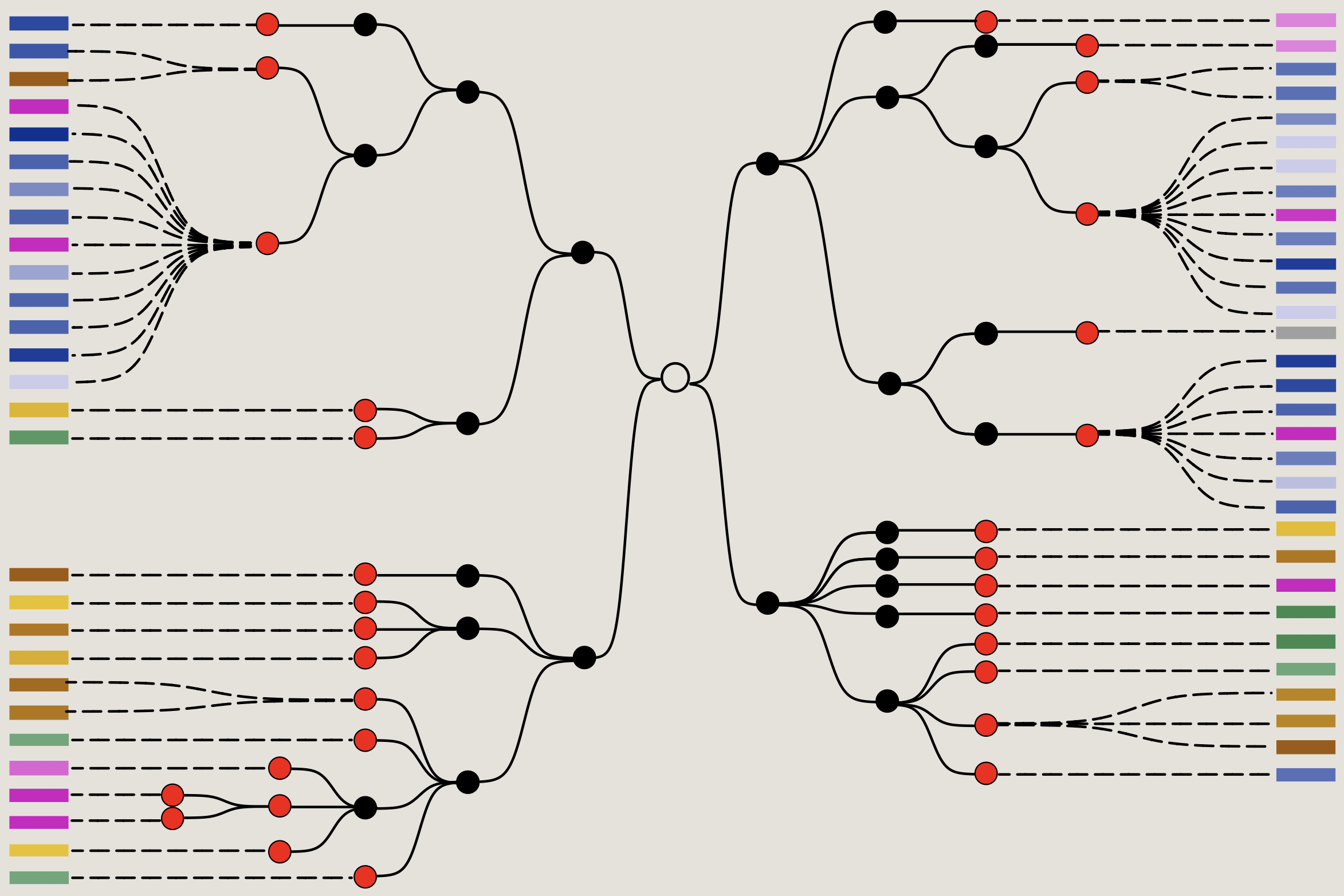
We developed a technology that adapts the CRISPR-Cas9 toolbox to infer cell lineage histories using mutations encoded in a genomic barcode.
In our method, scGESTALT, diverse, irreversible, Cas9-induced edits accumulate over many rounds of cell division, and the patterns of shared edits are used to reconstruct cell lineages. This is coupled with single-cell transcriptomics to generate single-cell resolved brain lineage trees.
NEXT GEN RECORDERS
We are exploring innovative ways to apply CRISPR-Cas genome editing for generating tools that query additional developmental paradigms.
These novel barcoding methods will address fundamental questions of how multiple biological pathways converge to regulate neuronal cell fates. Get in touch to learn more about this exciting new chapter in the lab